Determinants of Antigenicity in Tumor Neoepitopes for the Development of Personalized/Multiple Peptide Vaccines
Grant Keller, Department of Chemistry & Biochemistry and Harper Cancer Research Institute, University of Notre Dame; Hakimeh Ebrahimi-Nik, Department of Immunology, University of Connecticut School of Medicine; Lauren Davancaze, Department of Chemistry & Biochemistry and Harper Cancer Research Institute, University of Notre Dame; Alyssa Arbuiso, Department of Chemistry & Biochemistry and Harper Cancer Research Institute, University of Notre Dame; Pramod K Srivastava, Department of Immunology, University of Connecticut School of Medicine; Brian M. Baker, Department of Chemistry & Biochemistry and Harper Cancer Research Institute, University of Notre Dame
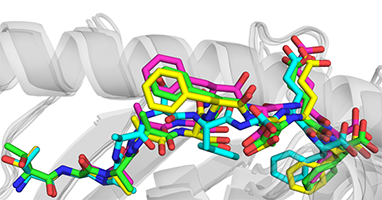
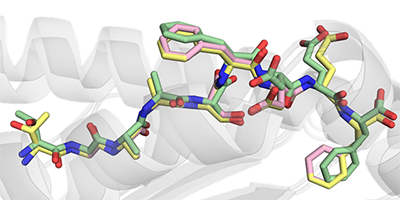
Immunotherapies present a promising approach for targeted treatment of cancer. Peptide vaccines incorporating mutant tumor peptides, or “neoantigens,” represent a low-cost and broadly applicable type of immunotherapy which can sensitize the immune system to tumors, overcome tolerance, and confer long-lasting immunologic memory with fine specificity. However, predicting which candidate neoantigens will likely be immunogenic is difficult and imprecise, which limits the applicability of peptide vaccines in spontaneous tumors. Previously we showed that structurally-encoded features of peptide-MHC proteins can be used to predicted immunogenicity better than many existing tools, but this method is still in its infancy and has only been tested on HLA-A*02:01-restricted nonameric peptides. In this study, we identified a group of mutant peptides presented by class I MHC in a non-spontaneous murine cancer model and rationalized the observed immunogenicity with molecular modeling experiments, as predicted IC50 alone did not explain reactivity. After solving the crystal structure of one pair of neoantigens, we propose ways to improve peptide-MHC model selection.